To make the working stock of the primers.
- 90 ul of Nuclease free (or Nanopure) H20 to 1.5 ml tube
- 10 ul of Stock primers
- Vortex briefly
I made these carefully in order to produce the following 12 pairs of primers.
| 1626 | HSP70c_FWD | AGGAAAGGTCGGGAGAGGAA | JH | 5/21/2015 | 20 | 55 | O.lurida | Heat shock 70 kDa protein 12A | |
| 1625 | HSP70c_REV | ACCTCGGACTTTGGACGAAC | JH | 5/21/2015 | 20 | 55 | O.lurida | Heat shock 70 kDa protein 12A | |
| 1624 | p29ING4_FWD | TACCTTTGGGCTTCACCGTC | JH | 5/21/2015 | 20 | 55 | O.lurida | Inhibitor of growth protein 4 (p29ING4) | |
| 1623 | p29ING4_REV | GTCCATCACACACCCCTCAG | JH | 5/21/2015 | 20 | 55 | O.lurida | Inhibitor of growth protein 4 (p29ING4) | |
| 1622 | CerS2_FWD | TTGTCGGTCTCCTCCTGCTA | JH | 5/21/2015 | 20 | 55 | O.lurida | Ceramide synthase 2 (CerS2) (LAG1 longevity assurance homolog 2) | |
| 1621 | CerS2_REV | CCGTCTTCTGAGCCATCGTT | JH | 5/21/2015 | 20 | 55 | O.lurida | Ceramide synthase 2 (CerS2) (LAG1 longevity assurance homolog 2) | |
| 1620 | GABABR1_FWD | CCGAGGAGGACACGAAACTC | JH | 5/21/2015 | 20 | 55 | O.lurida | Gamma-aminobutyric acid type B receptor subunit 1 (GABA-B receptor 1) (GABA-B-R1) (GABA-BR1) (GABABR1) (Gb1) | |
| 1619 | GABABR1_REV | CGGACAGGTTCTGGATTCCG | JH | 5/21/2015 | 20 | 55 | O.lurida | Gamma-aminobutyric acid type B receptor subunit 1 (GABA-B receptor 1) (GABA-B-R1) (GABA-BR1) (GABABR1) (Gb1) | |
| 1618 | HSP70d_FWD | TTTGTCTCACCGGCTTTGTG | JH | 5/21/2015 | 20 | 55 | O.lurida | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B') | |
| 1617 | HSP70d_REV | GACATGAGACCAAAGACGCC | JH | 5/21/2015 | 20 | 55 | O.lurida | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B') | |
| 1616 | THRa_FWD | GACACTATCCTCACTCGGCG | JH | 5/21/2015 | 20 | 55 | O.lurida | Thyroid hormone receptor alpha (Nuclear receptor subfamily 1 group A member 1) | |
| 1615 | THRa_REV | GGGTGCCGAGTAAACAAGGA | JH | 5/21/2015 | 20 | 55 | O.lurida | Thyroid hormone receptor alpha (Nuclear receptor subfamily 1 group A member 1) | |
| 1614 | Defensin_FWD | TCTAGCGGAGTTTGTTGGGG | JH | 5/21/2015 | 20 | 55 | O.lurida | Big defensin | |
| 1613 | Defensin_REV | ATGGCTGTCGGAGGAGGATT | JH | 5/21/2015 | 20 | 55 | O.lurida | Big defensin | |
| 1612 | GRB2_FWD | AACTTTGTCCACCCAGACGG | JH | 5/21/2015 | 20 | 55 | O.lurida | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (Protein Ash) (SH2/SH3 adapter GRB2) | |
| 1611 | GRB2_REV | CCAGTTGCAGTCCACTTCCT | JH | 5/21/2015 | 20 | 55 | O.lurida | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (Protein Ash) (SH2/SH3 adapter GRB2) | |
| 1610 | H3.3_FWD | CACGCTCTCCTCGAATCCTC | JH | 5/21/2015 | 20 | 55 | O.lurida | Histone H3.3 | |
| 1609 | H3.3_REV | AAGTTGCCTTTCCAGCGTCT | JH | 5/21/2015 | 20 | 55 | O.lurida | Histone H3.3 | |
| 1608 | H2A.V_FWD | TGCTTTCTGTGTGCCCTTCT | JH | 5/21/2015 | 20 | 55 | O.lurida | Histone H2A.V (H2A.F/Z) (Fragment) | |
| 1607 | H2A.V_REV | TATCACACCCCGTCACTTGC | JH | 5/21/2015 | 20 | 55 | O.lurida | Histone H2A.V (H2A.F/Z) (Fragment) | |
| 1606 | H2A_FWD | GCTGGGGTTTTTCTGGGTCT | JH | 5/21/2015 | 20 | 55 | O.lurida | Histone H2A | |
| 1605 | H2A_REV | GGAACTACGCCGAGAGAGTG | JH | 5/21/2015 | 20 | 55 | O.lurida | Histone H2A |
After producing the working stocks I then made a 10 reaction master mix.
Master Mix reagent table
| Volume | Reactions X10 | |
| Ssofast Evagreen MM | 10 | 100 |
| FWD Primer | 0.5 | 5 |
| REV Primer | 0.5 | 5 |
| Nanopure H2O | 8 | 80 |
| cDNA | 1 |
Protocol:
- Added Ssofast to each of 12 tubes
- Added Nanopure water to each of 12 tubes
- Carefully added FWD primer, then Reverse primer to the appropriate tube
- Vortex briefly
- One Master Mix used per column in plate
- Pipette 19 ul of master mix to each well for the appropriate column
- Either No Template Control (NTC) or sample for each Row in the plate
- 1 ul Sample/NTC per well in the appropriate row
Table Layout:
| HSP70c | P29ING | CerS2 | GABABR | HSP70d | THRa | Defensin | GRB2 | H3.3 | H2A.V | H2A | |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC | |
| NT1 | NT1 | NT1 | NT1 | NT1 | NT1 | NT1 | NT1 | NT1 | NT1 | NT1 | |
| HT1 | HT1 | HT1 | HT1 | HT1 | HT1 | HT1 | HT1 | HT1 | HT1 | HT1 | |
| ST1 | ST1 | ST1 | ST1 | ST1 | ST1 | ST1 | ST1 | ST1 | ST1 | ST1 | |
| NC1 | NC1 | NC1 | NC1 | NC1 | NC1 | NC1 | NC1 | NC1 | NC1 | NC1 | |
| HC1 | HC1 | HC1 | HC1 | HC1 | HC1 | HC1 | HC1 | HC1 | HC1 | HC1 | |
| SC1 | SC1 | SC1 | SC1 | SC1 | SC1 | SC1 | SC1 | SC1 | SC1 | SC1 | |
| NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC | NTC |
qPCR Program:
| Sybr New Plate+Sybr cDNA 60 melt 2 Read | ||
| Step | Temperature | Time |
| Initiation | 95 C | 10 min |
| Elongation | 95 C | 30 sec |
| 55 C | 1 min | |
| Read | ||
| 72 C | 30 sec | |
| Read | ||
| Repeat Elongation 39 times | ||
| Termination | 95 C | 1 min |
| 55 C | 1 sec | |
| Melt Curve Manual ramp 0.2C per sec Read 0.5 C | 55 - 95 C | 30 sec |
| 21 C | 10 min | |
| End |
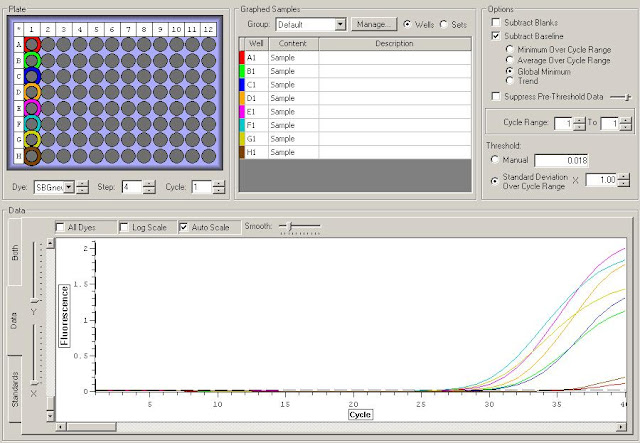
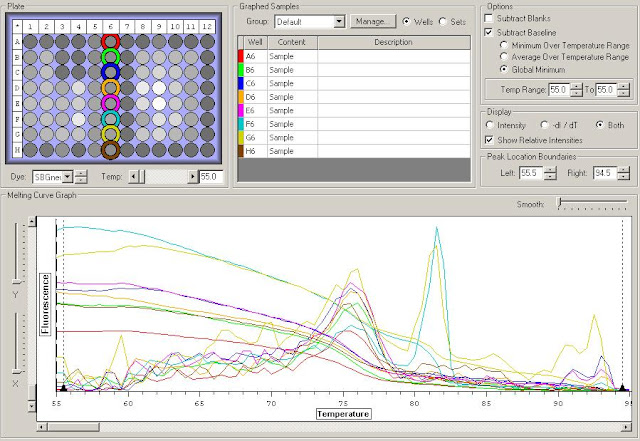
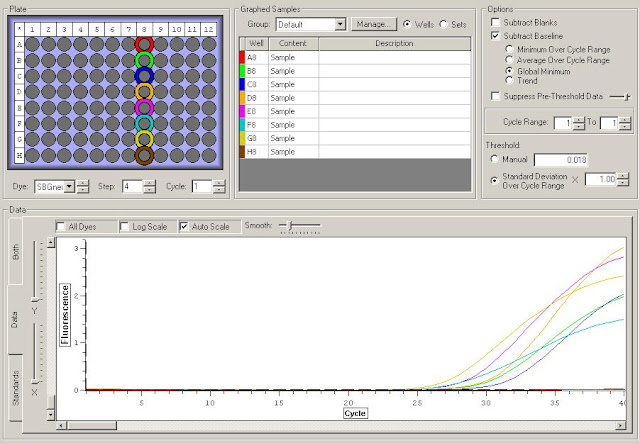
Results:
HSP70c Amplification
HSP70c Melt Curve
p29ING Amplification
p29ING Melt Curve
CerS2 Amplification
CerS2 Melt Curve
GABABR Amplification
GABABR Melt Curve
HSP70d Amplification
HSP70d Melt Curve
THRa Amplification
THRa Melt Curve
Defensin Amplification
Defensin Melt Curve
GRB2 Amplification
GRB2 Melt Curve
H3.3 Amplification
H3.3 Melt Curve
H2A.V Amplification
H2A.V Melt Curve
H2A Amplification
H2A Melt Curve
Posted below are assumptions based on the very limited data from this check. It is only being produced to eliminate useless primers from future tests and highlight possibly interesting primers. Only with further testing will we be able to determine true trends.
CerS2, HSP70d, THRa, and Defensin failed due to no amplification or poor melt curve conditions.
HSP70c produced a weird bump inline with the other melt curves. This primer should probably not be used due to the inability to eliminate this issue.
H3.3, H2A.V, and H2A all showed similar results with H2A being very similar in all samples. These could be candidates for normalizing genes to use with Actin.
p29ING also appeared to have uniform expression in all samples. This could also be another normalizing gene.
GABABR appeared to have down regulation in Heat Stress for Fidalgo and Dabob but there's no way to tell unless we run more replicates.
GRB2 had some variable expression between heat shock and control samples but we'll need to run further tests to ensure this.
Tomorrow I should finalize a list of the primers of interest. Hopefully next week I'll be able to run them with a full complement of samples and normalizing genes.
You can find the raw qPCR data here.



















No comments:
Post a Comment