Primers:
| 1620 | GABABR1_FWD | CCGAGGAGGACACGAAACTC | JH | 5/21/2015 | 20 | 55 | O.lurida | Gamma-aminobutyric acid type B receptor subunit 1 (GABA-B receptor 1) (GABA-B-R1) (GABA-BR1) (GABABR1) (Gb1) | Q9WV18 | |
| 1619 | GABABR1_REV | CGGACAGGTTCTGGATTCCG | JH | 5/21/2015 | 20 | 55 | O.lurida | Gamma-aminobutyric acid type B receptor subunit 1 (GABA-B receptor 1) (GABA-B-R1) (GABA-BR1) (GABABR1) (Gb1) | Q9WV18 |
Reagent Table:
| Volume | Reactions X116 | |
| Ssofast Evagreen MM | 10 | 1160 |
| FWD Primer | 0.5 | 58 |
| REV Primer | 0.5 | 58 |
| 1:9 cDNA | 9 |
- Added reagents from greatest to least volume
- Vortexed
- Centrifuged briefly
- Pipetted 11 ul Master Mix to each tube
- Pipetted 9 ul of 1:9 cDNA each column using a channel pipetter
- Centrifuged plate at 2000 rpm for 1 minute
- Ran Program Below
Program:
| Step | Temperature | Time |
| Initiation | 95 C | 10 min |
| Elongation | 95 C | 30 sec |
| 60 C | 1 min | |
| Read | ||
| 72 C | 30 sec | |
| Read | ||
| Repeat Elongation 39 times | ||
| Termination | 95 C | 1 min |
| 55 C | 1 sec | |
| Melt Curve Manual ramp 0.2C per sec Read 0.5 C | 55 - 95 C | 30 sec |
| 21 C | 10 min | |
| End |
Plate Layout:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| DNased 42215 HC1 | DNased 42215 NC1 | DNased 42215 SC1 | DNased 42215 HT1 1 | DNased 42215 NT1 1 | DNased 42215 ST1 1 | NTC |
| DNased 42215 HC2 | DNased 42215 NC2 | DNased 42215 SC2 | DNased 42215 HT1 2 | DNased 42215 NT1 2 | DNased 42215 ST1 2 | NTC |
| DNased 42215 HC3 | DNased 42215 NC3 | DNased 42215 SC3 | DNased 42215 HT1 3 | DNased 42215 NT1 3 | DNased 42215 ST1 3 | NTC |
| DNased 42215 HC4 | DNased 42215 NC4 | DNased 42215 SC4 | DNased 42215 HT1 4 | DNased 42215 NT1 4 | DNased 42215 ST1 4 | NTC |
| DNased 42215 HC5 | DNased 42215 NC5 | DNased 42215 SC5 | DNased 42215 HT1 5 | DNased 42215 NT1 5 | DNased 42215 ST1 5 | |
| DNased 42215 HC6 | DNased 42215 NC6 | DNased 42215 SC6 | DNased 42215 HT1 6 | DNased 42215 NT1 6 | DNased 42215 ST1 6 | |
| DNased 42215 HC7 | DNased 42215 NC7 | DNased 42215 SC7 | DNased 42215 HT1 7 | DNased 42215 NT1 7 | DNased 42215 ST1 7 | |
| DNased 42215 HC8 | DNased 42215 NC8 | DNased 42215 SC8 | DNased 42215 HT1 8 | DNased 42215 NT1 8 | DNased 42215 ST1 8 |
Results:
All samples
NTCs
The amplification curves look great in this one. There is no amplification in the NTCs which is perfect. To better analyse the data I ran the raw fluorescence through my script to produce adjusted expression bar graph, Two Way ANOVA, One Way ANOVA, Tukey's Test, and T-Test.
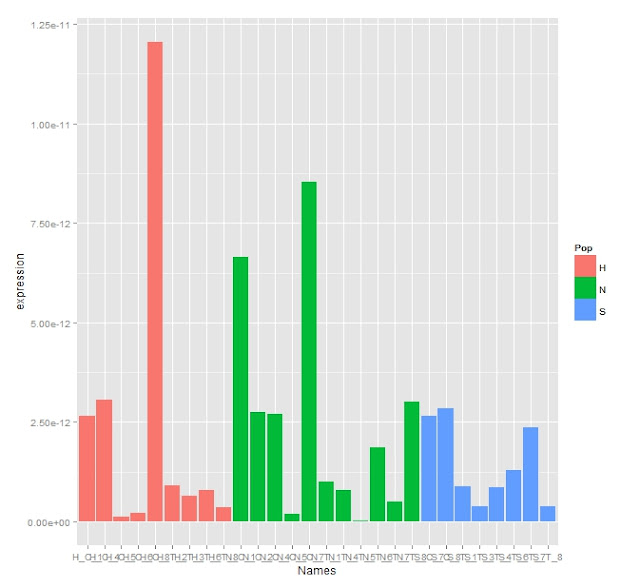
Adjusted Expression Graph
TWO WAY ANOVA with Tukey's TEST
Call:
aov(formula = expression ~ Pop + Treat + Pop:Treat, data = rep2res2)
Terms:
Pop Treat Pop:Treat Residuals
Sum of Squares 5.831470e-24 4.605870e-23 1.717210e-24 1.500405e-22
Deg. of Freedom 2 1 2 22
Residual standard error: 2.611517e-12
Estimated effects may be unbalanced
> TukeyHSD(fit)
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = expression ~ Pop + Treat + Pop:Treat, data = rep2res2)
$Pop
diff lwr upr p adj
N-H 2.366774e-13 -2.711956e-12 3.185311e-12 0.9778584
S-H -8.557194e-13 -4.043450e-12 2.332011e-12 0.7806364
S-N -1.092397e-12 -4.140705e-12 1.955911e-12 0.6458397
$Treat
diff lwr upr p adj
T-C -2.51348e-12 -4.581732e-12 -4.452286e-13 0.0194822
$`Pop:Treat`
diff lwr upr p adj
N:C-H:C 5.477504e-13 -4.597423e-12 5.692924e-12 0.9993875
S:C-H:C -8.619841e-13 -7.668409e-12 5.944440e-12 0.9985849
H:T-H:C -2.942336e-12 -8.399616e-12 2.514944e-12 0.5583572
N:T-H:C -2.420009e-12 -7.346137e-12 2.506119e-12 0.6494947
S:T-H:C -2.597238e-12 -7.523366e-12 2.328891e-12 0.5810595
S:C-N:C -1.409734e-12 -8.216159e-12 5.396690e-12 0.9860417
H:T-N:C -3.490086e-12 -8.947367e-12 1.967194e-12 0.3777506
N:T-N:C -2.967759e-12 -7.893888e-12 1.958369e-12 0.4412795
S:T-N:C -3.144988e-12 -8.071116e-12 1.781140e-12 0.3795617
H:T-S:C -2.080352e-12 -9.125670e-12 4.964967e-12 0.9371575
N:T-S:C -1.558025e-12 -8.200415e-12 5.084365e-12 0.9758792
S:T-S:C -1.735254e-12 -8.377644e-12 4.907137e-12 0.9618634
N:T-H:T 5.223269e-13 -4.728944e-12 5.773597e-12 0.9995598
S:T-H:T 3.450983e-13 -4.906172e-12 5.596369e-12 0.9999425
S:T-N:T -1.772287e-13 -4.874108e-12 4.519650e-12 0.9999963
ONE WAY ANOVA comparing POPULATIONS CONTROL
> fit2<-aov(expression~Pop, data=rep2res2[Treat=="C"])
> fit2
Call:
aov(formula = expression ~ Pop, data = rep2res2[Treat == "C"])
Terms:
Pop Residuals
Sum of Squares 2.90037e-24 1.41338e-22
Deg. of Freedom 2 9
Residual standard error: 3.962855e-12
Estimated effects may be unbalanced
> TukeyHSD(fit2)
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = expression ~ Pop, data = rep2res2[Treat == "C"])
$Pop
diff lwr upr p adj
N-H 5.477504e-13 -6.449936e-12 7.545437e-12 0.9740886
S-H -8.619841e-13 -1.011905e-11 8.395085e-12 0.9635753
S-N -1.409734e-12 -1.066680e-11 7.847334e-12 0.9061601
ONE WAY ANOVA comparing POPULATIONS TREATMENT
> fit3<-aov(expression~Pop, data=rep2res2[Treat=="T"])
> fit3
Call:
aov(formula = expression ~ Pop, data = rep2res2[Treat == "T"])
Terms:
Pop Residuals
Sum of Squares 6.585500e-25 8.702475e-24
Deg. of Freedom 2 13
Residual standard error: 8.181816e-13
Estimated effects may be unbalanced
> TukeyHSD(fit3)
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = expression ~ Pop, data = rep2res2[Treat == "T"])
$Pop
diff lwr upr p adj
N-H 5.223269e-13 -8.721776e-13 1.916831e-12 0.5963155
S-H 3.450983e-13 -1.049406e-12 1.739603e-12 0.7937301
S-N -1.772287e-13 -1.424511e-12 1.070054e-12 0.9257775
T-TEST FOR DABOB
> fit4<-t.test(expression~Treat, data=rep2res2[Pop=="H"])
> fit4
Welch Two Sample t-test
data: expression by Treat
t = 1.3399, df = 4.024, p-value = 0.2509
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-3.140394e-12 9.025065e-12
sample estimates:
mean in group C mean in group T
3.618621e-12 6.762853e-13
T-TEST FOR FIDALGO
> fit5<-t.test(expression~Treat, data=rep2res2[Pop=="N"])
> fit5
Welch Two Sample t-test
data: expression by Treat
t = 1.8959, df = 4.684, p-value = 0.1204
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-1.139381e-12 7.074900e-12
sample estimates:
mean in group C mean in group T
4.166372e-12 1.198612e-12
T-TEST FOR OYSTER BAY
> fit6<-t.test(expression~Treat, data=rep2res2[Pop=="S"])
> fit6
Welch Two Sample t-test
data: expression by Treat
t = 5.4509, df = 5.775, p-value = 0.001794
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
9.488834e-13 2.521624e-12
sample estimates:
mean in group C mean in group T
2.756637e-12 1.021384e-12
The stats show there are no differences between populations but that there is a significant difference between the treatment and control. The T-TESTs show that this difference lies in the Oyster Bay Population which is different between treatment and control. This is apparent when looking at the boxplot. Though looking at the data set it appears that its only based on an n of 5 which severely limits the strength of this data.
You can see the raw data set here.



No comments:
Post a Comment