Primers:
| 1682 | EF1d_FWD | GAACTGCCCACTGATTTGCC | JH | 7/22/2015 | 20 | 55 | 60 | O.lurida | Elongation factor 1-delta (EF-1-delta) | A5D989 |
| 1681 | EF1d_REV | TGTGGGGTGAAACACGTTGA | JH | 7/22/2015 | 20 | 50 | 60 | O.lurida | Elongation factor 1-delta (EF-1-delta) | A5D989 |
| Volume | Reactions X116 | |
| Ssofast Evagreen MM | 10 | 1160 |
| FWD Primer | 0.5 | 58 |
| REV Primer | 0.5 | 58 |
| 1:9 cDNA | 9 |
- Added reagents from greatest to least volume
- Vortexed
- Centrifuged briefly
- Pipetted 11 ul Master Mix to each tube
- Pipetted 9 ul of 1:9 cDNA each column using a channel pipetter
- Centrifuged plate at 2000 rpm for 1 minute
- Ran Program Below
Program:
| Step | Temperature | Time |
| Initiation | 95 C | 10 min |
| Elongation | 95 C | 30 sec |
| 60 C | 1 min | |
| Read | ||
| 72 C | 30 sec | |
| Read | ||
| Repeat Elongation 39 times | ||
| Termination | 95 C | 1 min |
| 55 C | 1 sec | |
| Melt Curve Manual ramp 0.2C per sec Read 0.5 C | 55 - 95 C | 30 sec |
| 21 C | 10 min | |
| End |
Plate Layout:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| DNased 42215 HC1 | DNased 42215 NC1 | DNased 42215 SC1 | DNased 42215 HT1 1 | DNased 42215 NT1 1 | DNased 42215 ST1 1 | NTC |
| DNased 42215 HC2 | DNased 42215 NC2 | DNased 42215 SC2 | DNased 42215 HT1 2 | DNased 42215 NT1 2 | DNased 42215 ST1 2 | NTC |
| DNased 42215 HC3 | DNased 42215 NC3 | DNased 42215 SC3 | DNased 42215 HT1 3 | DNased 42215 NT1 3 | DNased 42215 ST1 3 | NTC |
| DNased 42215 HC4 | DNased 42215 NC4 | DNased 42215 SC4 | DNased 42215 HT1 4 | DNased 42215 NT1 4 | DNased 42215 ST1 4 | NTC |
| DNased 42215 HC5 | DNased 42215 NC5 | DNased 42215 SC5 | DNased 42215 HT1 5 | DNased 42215 NT1 5 | DNased 42215 ST1 5 | |
| DNased 42215 HC6 | DNased 42215 NC6 | DNased 42215 SC6 | DNased 42215 HT1 6 | DNased 42215 NT1 6 | DNased 42215 ST1 6 | |
| DNased 42215 HC7 | DNased 42215 NC7 | DNased 42215 SC7 | DNased 42215 HT1 7 | DNased 42215 NT1 7 | DNased 42215 ST1 7 | |
| DNased 42215 HC8 | DNased 42215 NC8 | DNased 42215 SC8 | DNased 42215 HT1 8 | DNased 42215 NT1 8 | DNased 42215 ST1 8 |
Results:
Opticon
All
Amp
Melt Curve
NTCs
Amp
Melt Curve
CFX
All
NTCs
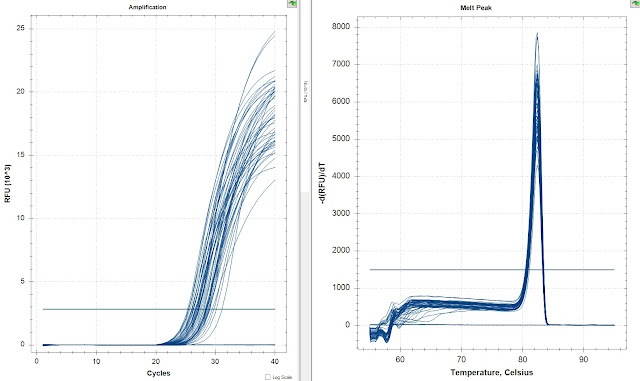
The amplification curves in both runs look great. There's no amplification in either set of NTCs. Also the latest amplifying samples were they same in both replicates. I then compared the replicates with the Opticon Correction to see how close are they.
There is less than a 2 ct difference between the replicates with correction. The there is one major outlier which is likely caused by mechanical error.
I will tomorrow I will average the Ct values and efficiencies to develop data to normalize expression data for the other targets. Also I've been reading on how to deal with outliers. If I throw out the data, then it screws up the statistics. One way I've seen this dealt with in the literature is to assume that outliers are caused by mechanical/human error and replace the values with the latest Ct value and efficiency found in the data which assumes the lowest observed expression.
You can see paper here:
Differential gene expression profiling of Listeria monocytogenes in Cacciatore and Felino salami to reveal potential stress resistance biomarkers
http://www.sciencedirect.com/science/article/pii/S0740002014002329